 Olfaction offers a wealth of biological problems awaiting understanding in molecular terms. We’re interested in the remarkable sensitivity of the olfactory system, its ability to distinguish among odors, and the mechanisms by which sensory input is translated into behavioral output. Drosophila has a highly sophisticated olfactory system, which we study with molecular, genetic, physiological, computational, and behavioral approaches.
Olfaction offers a wealth of biological problems awaiting understanding in molecular terms. We’re interested in the remarkable sensitivity of the olfactory system, its ability to distinguish among odors, and the mechanisms by which sensory input is translated into behavioral output. Drosophila has a highly sophisticated olfactory system, which we study with molecular, genetic, physiological, computational, and behavioral approaches.
We discovered a family of 60 seven-transmembrane-domain genes, the Or genes, which encode odor receptors. We identified them by using a novel computer algorithm to search the fly genome database for proteins with a particular structure, as opposed to proteins with a particular sequence. Different receptors are expressed in different subsets of olfactory receptor neurons.
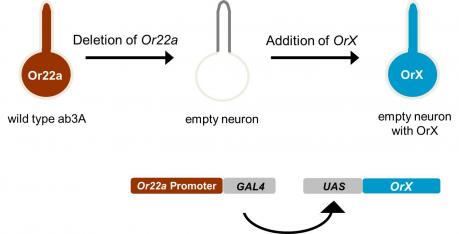 We have examined the odor receptors using an in vivo expression system that we call the “empty neuron” system. The system is based on a mutant antennal neuron, the empty neuron, that lacks endogenous odorant receptors and that does not respond to odors. Receptors are systematically expressed in the empty neuron, and we measure the odorant responses that each individual receptor confers. In this way we are able to analyze how odor quality, quantity, and duration are encoded by a receptor repertoire.
We have examined the odor receptors using an in vivo expression system that we call the “empty neuron” system. The system is based on a mutant antennal neuron, the empty neuron, that lacks endogenous odorant receptors and that does not respond to odors. Receptors are systematically expressed in the empty neuron, and we measure the odorant responses that each individual receptor confers. In this way we are able to analyze how odor quality, quantity, and duration are encoded by a receptor repertoire.
![]() In an effort to predict the perceptual quality of odors we constructed a 21-dimensional odor space from the responses of the 21 larval receptors: each dimension represents the response of one receptor, and each odor maps to a point in the space. We found that the distance between odors in the space correlates with the extent to which one odor masks the other. If odors A and B map near each other in the space, then a background of odor A tends to block the response to odor B.
In an effort to predict the perceptual quality of odors we constructed a 21-dimensional odor space from the responses of the 21 larval receptors: each dimension represents the response of one receptor, and each odor maps to a point in the space. We found that the distance between odors in the space correlates with the extent to which one odor masks the other. If odors A and B map near each other in the space, then a background of odor A tends to block the response to odor B.
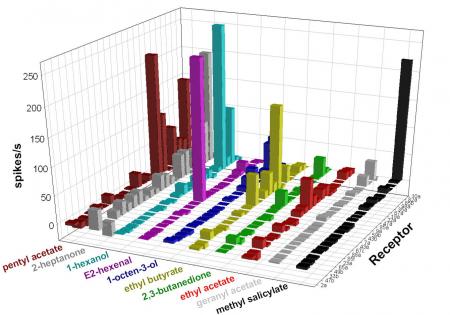
We’re interested in the circuitry through which olfactory input is translated into olfactory responses. Mutational analysis has shown that different receptors mediate responses to different concentrations of an odorant, and we’ve found that the activity of a small number of receptors is a powerful predictor of behavior, but the mechanisms that drive olfactory behavior are largely unknown. We’re currently investigating how the olfactory circuit is activated, how its activity is shaped by non-neuronal cells, and how its activity is integrated with that of other circuits.
We recently discovered exitrons (exonic introns) in four Odorant receptor (Or) genes of Drosophila. Exitrons are sequences that can be spliced from within a protein-coding exon, thereby altering the encoded protein. We are unaware of previous reports of exitrons in Drosophila. Removal of the exitron from the pheromone receptor gene Or88a creates a non-coding transcript. The exitron has been conserved for 20 million years. Activation of Or88a- expressing olfactory receptor neurons via either optogenetics or pheromone stimulation increased the level of these non-coding transcripts, suggesting that exitron splicing could act in neuronal modulation.
We recently generated an atlas of linear and circular lncRNAs in the Drosophila olfactory system. They create an lncRNA-to-neuron map, which shows that olfactory receptor neurons are defined not only by their odor receptors but also by the combination of lncRNAs that they express. Interestingly, a number of species-specific lncRNAs are enriched in pheromone-sensing neurons.
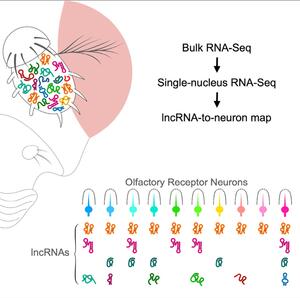
A lncRNA-to-neuron map
References
Shang, X., Talross, G.J.S., and Carlson, J.R. (2024) Exitron splicing of odor receptor genes in Drosophila, PNAS 121 (13) e2320277121.
Talross, G.J.S and Carlson, J.R. (2023) The rich non-coding RNA landscape of the Drosophila antenna, Cell Reports 42, 112482, 1-19.
Xiao, S., Sun, J.S., and Carlson, J.R. (2019), Robust olfactory responses in the absence of odorant binding proteins, eLife, e51040.
Sun, J.S., Larter, N., Chahda, J.S., Rioux, D., Gumaste, A., and Carlson, J.R. (2018) Humidity response depends on the small soluble protein Obp59a in Drosophila, eLife 2018;e39249
Mathew, D., Martelli, C., Kelley-Swift, E., Brusalis, C., Gershow, M., Samuel, A., Emonet, T., and Carlson, J. (2013) Functional diversity among sensory receptors in a Drosophila olfactory circuit, Proc. Natl. Acad. Sci. 110(23):E2134-43.
Kreher, S., Kim, J., and Carlson, J. (2008) Translation of sensory input into behavioral output via an olfactory system. Neuron 59, 110-124.
van der Goes van Naters, W. and Carlson, J.R. (2007) Receptors and neurons for fly odors in Drosophila. Current Biology 17(7), 606-12.
Hallem, E., and Carlson, J.R. (2006) Odor coding by a receptor repertoire. Cell 125, 143-160.
Hallem, E., Ho, M., and Carlson, J. (2004) The Molecular Basis of Odor Coding in the Drosophila Antenna, Cell 117, 965-979.